- From: Alan Ruttenberg <alanruttenberg@gmail.com>
- Date: Thu, 31 May 2007 03:28:42 -0400
- To: Eric Neumann <eneumann@teranode.com>, wilbanks@creativecommons.org, Jonathan Rees <jar@mumble.net>, William Bug <William.Bug@drexelmed.edu>, public-semweb-lifesci hcls <public-semweb-lifesci@w3.org>, Rob Williams <s2g2@mycingular.blackberry.net>, MaryAnn Martone <maryann@ncmir.ucsd.edu>, Gully Burns <gully@usc.edu>, Karen Skinner <ks79x@nih.gov>
- Message-Id: <E2712E62-007E-436E-BE08-8FEB69C90E70@gmail.com>
> Is there an easy way to get the http GET string for these queries?
> I'd
> love to start a page where non-semweb people can click and get answers
> to them...
>
> jtw
>
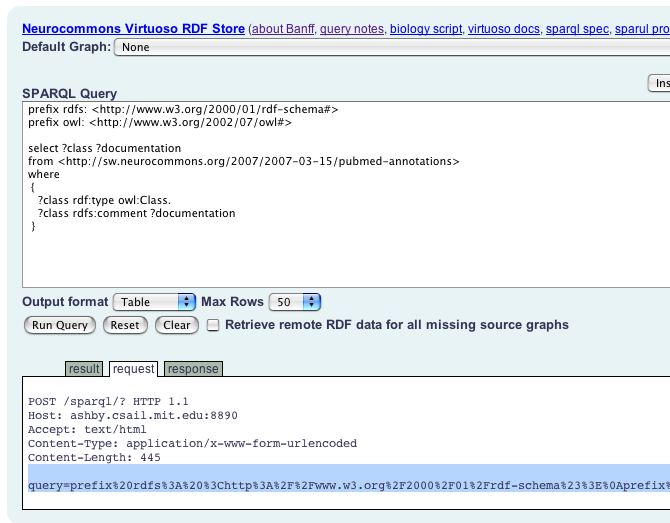
When you are using the sparql form and have a result, click on the
"request" tab. You'll see "query=..."
Append that to "http://hcls1.csail.mit.edu:8890/sparql/?"
Note that the form always has a limit. You may wish to adjust or
remove it - it is at the end of the query string, e.g.
query=prefix%20.....&maxrows=50
You can also look at the code in http://hcls1.csail.mit.edu:8890/map/
map.txt
In that case you use javascript to turn a plain text query into the
escaped form (if you copy/paste this note the query asks for results
returned as json, and for only a single result..
var plaintextquery =
"prefix sc: <http://sw.neurocommons.org/2007/annotations#> \
prefix aba: <http://sw.neurocommons.org/2007/aba/> \
prefix xsd: <http://www.w3.org/2001/XMLSchema#> \
SELECT ?url \
FROM <http://sw.neurocommons.org/aba-20070226> \
WHERE \
{ ?b rdf:type aba:MouseBrainGeneExpressionSurvey. \
?b aba:measuresGeneIdentifiedWith ?g. \
?g aba:hasSymbol \"THE_GENE_GOES_HERE\"^^xsd:string. \
?b aba:hasSectionSeries ?ss. \
?ss aba:hasSection ?bs. \
?bs aba:atPosition ?pos . filter (xsd:integer(?pos) =
THE_POSITION_GOES_HERE) . \
?bs aba:hasSectionOrientation aba:SagittalOrientation. \
?bs aba:hasImagePyramids ?ps. \
?ps aba:hasImagePyramid ?p. \
?p rdf:type aba:ExpressionImagePyramid. \
?p aba:hasURLTemplate ?url. \
} \
LIMIT 1";
var sparqlQuery= "/sparql/?query="+escape(plaintextquery)
+"&format=json&maxrows=1";
-Alan
On May 30, 2007, at 2:20 PM, Eric Neumann wrote:
> > John, > > I do recall doing this with Eric P's help while developing some of > the sparql queries for the Banff demo. I agree, this is one way to > 'wrap' some basic queries and allow users to select for GO, MESH, > and anatomical terms to get useful results back in (not to mention > that the results could have specific views made for the query > templates ala Exhibit). > > I am hoping we'll have a simple way to select for classes of gene's > using a GO-term (localization and bio process) select-GUI... > > Eric > > > -----Original Message----- > From: public-semweb-lifesci-request@w3.org on behalf of John Wilbanks > Sent: Wed 5/30/2007 10:32 AM > To: Jonathan Rees > Cc: William Bug; public-semweb-lifesci hcls; Alan Ruttenberg; Rob > Williams; MaryAnn Martone; Gully Burns; Karen Skinner > Subject: Re: Adding addition resources to the accruing in the > NeuroCommons > > > Is there an easy way to get the http GET string for these queries? > I'd > love to start a page where non-semweb people can click and get answers > to them... > > jtw > > Jonathan Rees wrote: > > Thanks for the suggestions. We'll be looking at these once we dig > out > > from under all the things that we had put off until after Banff. Of > > course, if someone else would like to tackle them first, that > would be > > great. > > > > It would be especially interesting to have good examples of queries > > that one would make that cross some of these sources with the > ones we > > already have. > > > > Jonathan > > > > On 5/24/07, William Bug <William.Bug@drexelmed.edu> wrote: > > > >> Hi Jonathan, > >> > >> Alan had mentioned at today's W3C HCLS IG call that you are > looking to > >> expand the resources you track in the NeuroCommons RDF > repository. He > >> mentioned work to add resources such as the ATCC. > >> > >> I mentioned there were a collection of resources being funded by > the NIH > >> Blueprint funding initiative that I would also suggest should be on > >> the list > >> for inclusion. Alan suggested I pass these on to you. > >> > >> You can find these described at: > >> http://neuroscienceblueprint.nih.gov/ > >> > >> There are a few that Alan has already begun to work on (or > examine for > >> inclusion) - e.g., CCDB, GENSAT, etc.. This page lists several > more such > >> resources that provide broad-scoped value in the realm of > >> neuroinformatics. > >> I'd particularly point to those resources listed under: > >> Proteins & Genes (includes the burgeoning NIH Neuroscience > Microarray > >> Consortium that Kei is a part of): > >> http://neuroscienceblueprint.nih.gov/neuroscience_resources/ > gene_protein.htm > >> > >> Cells, Tissues, and DNA > >> http://neuroscienceblueprint.nih.gov/neuroscience_resources/ > cell_tissue.htm > >> > >> Imaging Tools: > >> http://neuroscienceblueprint.nih.gov/neuroscience_resources/ > imaging_tools.htm > >> > >> Neuroinformatics: > >> http://neuroscienceblueprint.nih.gov/neuroscience_resources/ > neuroinformatics.htm > >> > >> > >> If you need any assistance in sorting through these resources, > don't > >> hesitate to ask. It might also be worthwhile having a brief > NeuroCommons > >> Working Group Teleconference with folks such as Rob WIlliams, > Maryann > >> Martone, Gully Burns, etc. to review these resources and help > you put > >> a list > >> of priorities in place. > >> > >> Cheers, > >> Bill > >> > >> > >> > >> Bill Bug > >> Senior Research Analyst/Ontological Engineer > >> > >> Laboratory for Bioimaging & Anatomical Informatics > >> www.neuroterrain.org > >> Department of Neurobiology & Anatomy > >> Drexel University College of Medicine > >> 2900 Queen Lane > >> Philadelphia, PA 19129 > >> 215 991 8430 (ph) > >> 610 457 0443 (mobile) > >> 215 843 9367 (fax) > >> > >> > >> Please Note: I now have a new email - William.Bug@DrexelMed.edu > > -- > ----------------------------------- > John Wilbanks > Executive Director, Science Commons > ----------------------------------- > > > >
Attachments
- image/png attachment: Picture_1.png
Received on Thursday, 31 May 2007 07:36:01 UTC