- From: M. Scott Marshall <mscottmarshall@gmail.com>
- Date: Thu, 16 Jun 2011 16:18:50 +0200
- To: Claus Stie Kallesøe <CSH@lundbeck.com>, linkedlifedatapracticesnote@googlegroups.com, HCLS <public-semweb-lifesci@w3.org>
- Cc: Anja Jentzsch <anja@anjeve.de>
- Message-ID: <BANLkTi=3E2uyG4a7MkOG4JdPSw_4Fr-19Q@mail.gmail.com>
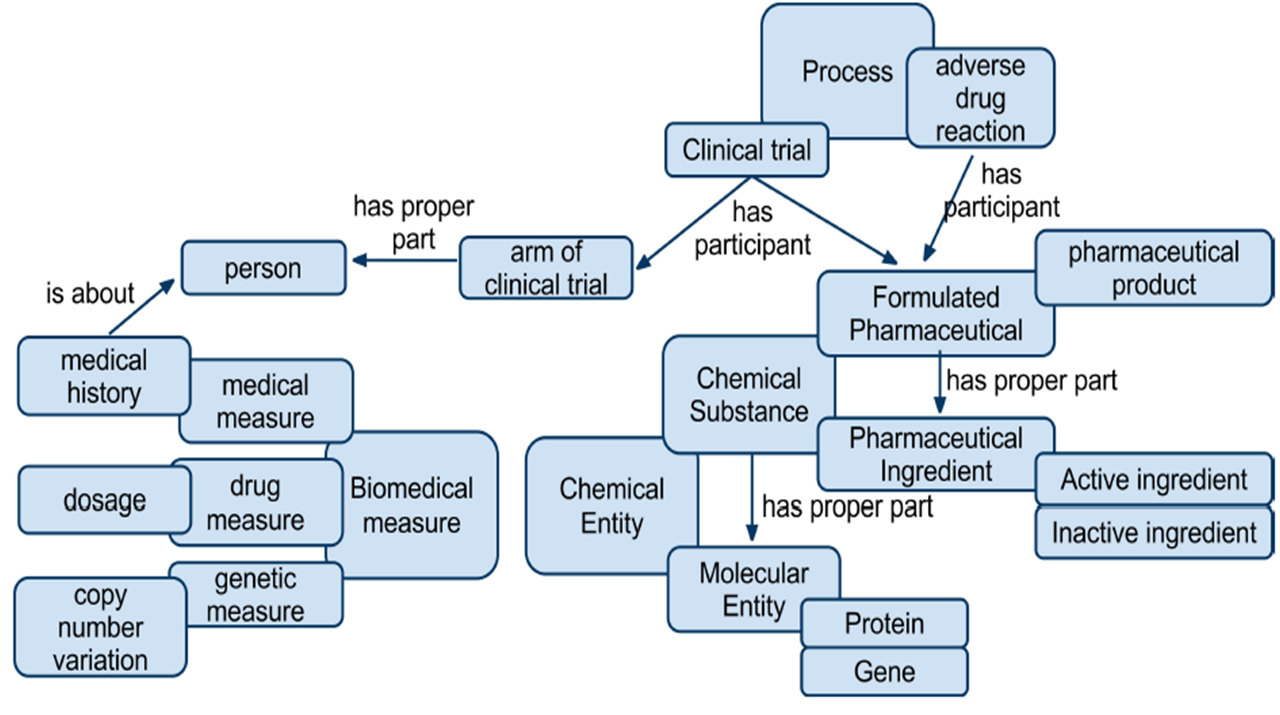
Hi Claus, [bouncing this to the list because I think this is of general interest and there is plenty of expertise there] This is where many get stuck. Happily, you are not the first one looking for specific classes/terms that match your data although there are plenty of areas of ontological usage that still need clarification and improvement. In TMO, we selected from a number of existing ontologies as much as we could and filled in where we couldn't find the right fit. See the attached image for an overview of the structure. TMO is also available from Bioportal. See also, the NCBO Ontology Recommender as a place to get started. http://www.bioontology.org/ontology-recommender http://www.bioontology.org/wiki/index.php/Ontology_Recommender_Client_Examples -Scott -- M. Scott Marshall, W3C HCLS IG co-chair, http://www.w3.org/blog/hcls http://staff.science.uva.nl/~marshall On Thu, Jun 16, 2011 at 3:35 PM, Claus Stie Kallesøe <CSH@lundbeck.com>wrote: > Ok, now I am ready. > > I will try to follow your steps so starting by creating an ontology for > LudnebckResearch (compound, batch, assay, project, scientist etc). > > I will use TopBraid composer. Its comprehensive (protege seems even harder) > but I think I have worked out what parts I need and from the neologism site > I found this - which is actually really helpful in order to understand what > it is and what it is not!: > > ---- > Neologism is no ontology editor. If you want to build large and complex > domain ontologies, if you need the full power of OWL, or if you need > sophisticated reasoning services, consider applications such as TopBraid > Composer, Protégé, or the NeOn Toolkit. Neologism is designed for the > simpler space of RDF vocabularies. OWL ontologies tend to be about modelling > a domain with the formal language of logics; RDF vocabularies thend to be > about exchanging and integrating data between systems and on the Web. > > ---- > > So when building an ontology i am thinking of something like > lundbeck:compound; lundbeck:scientist etc. As an overall research ontology. > I will then link these to the specifics in the relational databases like > lundbeck:compound SKOS:narrowmatch/OWL:sameAs ddis:compound (ddis being one > of the databases I have mapped with D2R). This is all the Phil approach. > > But as many of the concepts are known outside lundbeck (and we migth want > to link to that later) should I instead use something like this: > http://bioportal.bioontology.org/ontologies/45788 as "chemical entities of > biologicalinterest" is what I have? Or should I create my own and then later > do lundbeck:compound OWL:sameAS chebi:compound or similar? > > Remember the best practices paper several times mentioned always to > reuse.... > > Ok, my last question for today. > > claus > > > > -----Original Message----- > From: Anja Jentzsch [mailto:anja@anjeve.de] > Sent: Thursday, June 16, 2011 12:51 PM > To: Claus Stie Kallesøe > Cc: M. Scott Marshall > Subject: Re: Linkset Provenance > > > I had a look here: > > http://www4.wiwiss.fu-berlin.de/latc/toollibrary/example.php > > > > That might actually be some of the things I have been looking for. > > What tools to use for what, in what order and how to put it together. > > Anja, this also describes what to do with the link descriptions from > > Silk ;-) > > I wrote this tutorial and it is exactly what I described yesterday ;) > Probably I didn't put it specific enough. > > Anja > > > I will try to set something like this up to play > > > > c > > > > -----Original Message----- > > From: Anja Jentzsch [mailto:anja@anjeve.de] > > Sent: Wednesday, June 15, 2011 6:02 PM > > To: Claus Stie Kallesøe > > Cc: M. Scott Marshall > > Subject: Linkset Provenance > > > > I just put this on IRC but you're already gone: > > > > 17:00 Anja I wonder if it makes sense to have provenance data on the > linksets provided via D2R as a plugin as well > > 17:00 Anja this would cover the concerns Scott was talking about > > -- M. Scott Marshall, W3C HCLS IG co-chair, http://www.w3.org/blog/hcls http://staff.science.uva.nl/~marshall
Attachments
- image/png attachment: TMOStructure.png
Received on Thursday, 16 June 2011 14:19:20 UTC