- From: Michael Miller <Michael.Miller@systemsbiology.org>
- Date: Wed, 30 Nov 2011 12:13:03 -0800
- To: James Malone <malone@ebi.ac.uk>, HCLS <public-semweb-lifesci@w3.org>
- Cc: Marco Brandizi <brandizi@ebi.ac.uk>
- Message-ID: <e486913c3836fbb648647567c40cabb7@mail.gmail.com>
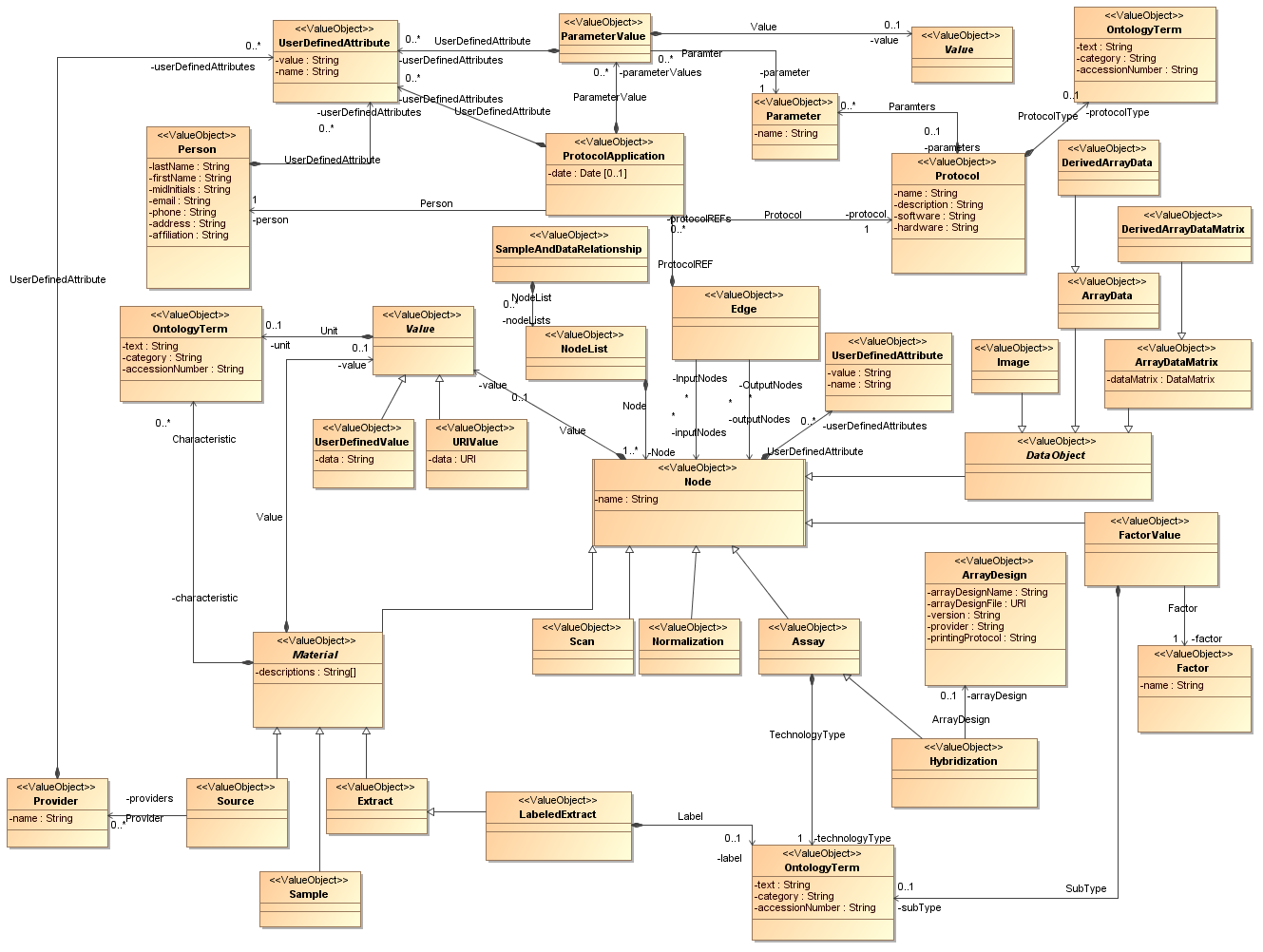
hi james, thanks for the example, it looks very promising. i believe this is jim's link to his illustration of his work (in no way reflects any effort on my part!): http://dl.dropbox.com/u/9752413/IRWG/carpt-00469.png not seeing the formal owl definition but just looking at the png, it looks like if the owl ontology was a realization of the MAGE-TAB object model (http://wwwdev.ebi.ac.uk/microarray-srv/magetab/generated-doc/objectmodel. html or ftp://smd-ftp.stanford.edu/smd/transfers/magetab/magetab.xml -- see attachments) a lot of the attributes and relations can be in a super term like Material and the realizable terms like Source, Sample, ..., can have 'is_a' or similar to OBI and EFO. in terms of making the site public, that would be great so that the W3C note could reference it and it could joined into LODD cheers, michael > -----Original Message----- > From: James Malone [mailto:malone@ebi.ac.uk] > Sent: Wednesday, November 30, 2011 9:23 AM > To: HCLS > Cc: Marco Brandizi > Subject: MAGE-TAB in RDF > > All, > > I had promised to send something on regarding the work our student > (Drashtti Vasant) had been doing on producing RDF from our MAGE-TAB > experiments in ArrayExpress. So here are some bits. We started this a > couple of months back before a model was sent around by Jim or Michael > (apologies, not sure who it was exactly - for whatever reason I can not > find the email and diagram that was sent round - could someone resend > it > please!). Interesting to see the things that align/different, I seem to > recall they aren't a million miles off. > > Anyway, I've attached the graphs we used as our baseline and the table > of nodes we included as rdf and the triples we formed in this rdf - > note > we do not include everything in the graphs attached. For factor values, > we ran our Zooma term matching across the values to try and mine these > against matches in EFO and for those that we matched we formed triples > (e.g. liver, cancer). For those we couldn't match automatically we > added > some default high level triples (which are less useful) just to say > this > has some factor values and we captured it as a literal text string. I'm > simplifying a bit as it was a bit more complicated but more or less > that > is what we did. As well as the triples in the doc I've attached we also > added some "convenience" triples to each of these nodes directly to the > experiment using part_of relation so you can do simple queries on the > experiment without having to traverse the whole graph. > > The store is sitting on an internal server at the moment but we may > open > it up if there is any interest in using it. Otherwise, we have some > internal (interesting) stuff we're doing with it as well expanding and > refining what we have. We focused on things we wanted to ask questions > about explicitly based on a set of competency questions we had formed. > It's not perfect but I'm of the school of release early, often, refine. > > Cheers, > > James > > PS credit for work is to Drashtti Vasant who did most of this > implementation, myself and Tony Burdett supervised with some guidance > on > RDF matters from Marco Brandizi. > > > -- > European Bioinformatics Institute, > Wellcome Trust Genome Campus, > Cambridge, CB10 1SD, > United Kingdom > Tel: + 44 (0) 1223 494 676 > Fax: + 44 (0) 1223 494 468
Attachments
Received on Wednesday, 30 November 2011 20:13:38 UTC