
Generated INChI:
If you have any question, please contact Yong Zhang(yz237@cam.ac.uk).
We show that the IUPAC Chemical Identifier (INChI) is a powerful and precise tool for indexing chemical structures both in databases and on the Web. We have developed Web services for the graphical creation of INChIs and submission as queries. The INChI is robust as a web-based query in the GoogleTM search engine and seems to have high recall and precision. We therefore recommend its suitability as an algorithmic approach to LSIDs (LifeScienceIdentifier) for small biological (organic) molecules. The findings also suggest that other LSIDs may be effectively indexed by web engines.
This is a supplement to our position paper to the W3C-SWLS activity suggesting the INChI as an LSID. It arises because of exciting results that have become available in the last week. If appropriate it can be demonstrated at the meeting, and instructions are given so that anyone with a browser can use it. We ask that it not be publicised widely until after the meeting and that any published use of it should cite this position paper.
For those not familiar with chemical search engines we should stress that up until INChI there was no simple way of searching for chemical structure on the web. Some systems use arbitrary proprietary IDs (such as the Chemical Abstracts Service registry number, "CAS number"). The CAS number - a 5-7 digit hyphenated string - is not open (it is, we believe, copyrighted by CAS) and on the web can retrieve many false positives such as football scores. For precise structure searching each server has to implement a bespoke search engine, usually proprietary, with no communality in query or behaviour. Moreover most of these only allow single graphical queries and are therefore unsuited to the Semantic Web. The SMILES language allows a canonical serialization of molecular structure, but is proprietary. Moreover several incompatible "unique SMILES" have been developed, making it impossible to use on the Web. It also does not behave well in search engines, giving many false positives (though to be fair, there is a much greater base of SMILES on the web).
We implemented an early version of the INChI software (V0.932beta) at http://wwmm.ch.cam.ac.uk. and generated INChIs for the 250K molecules in the National Cancer Institute dataset. We have used these INChIs as an internal index with apparently complete precision and recall. A small number of these molecules, along with their INChIs, were published on the Web, e.g. in discussion papers, examples, etc. There was initially no intention that these INChIs were other than human readable.
We have now discovered, serendipitously, that these INChIs have been comprehensively and accurately indexed by the Google search engine. From preliminary exploration it appears that every known document in which an INChI appears has been indexed and that all are retrievable by standard queries with virtually 100% precision. This means that standard Web-based indexers, without any alteration, are capable of acting as completely precise chemical search engines. Although we have many years of developing chemistry on the web, this was an unexpected and very welcome finding.
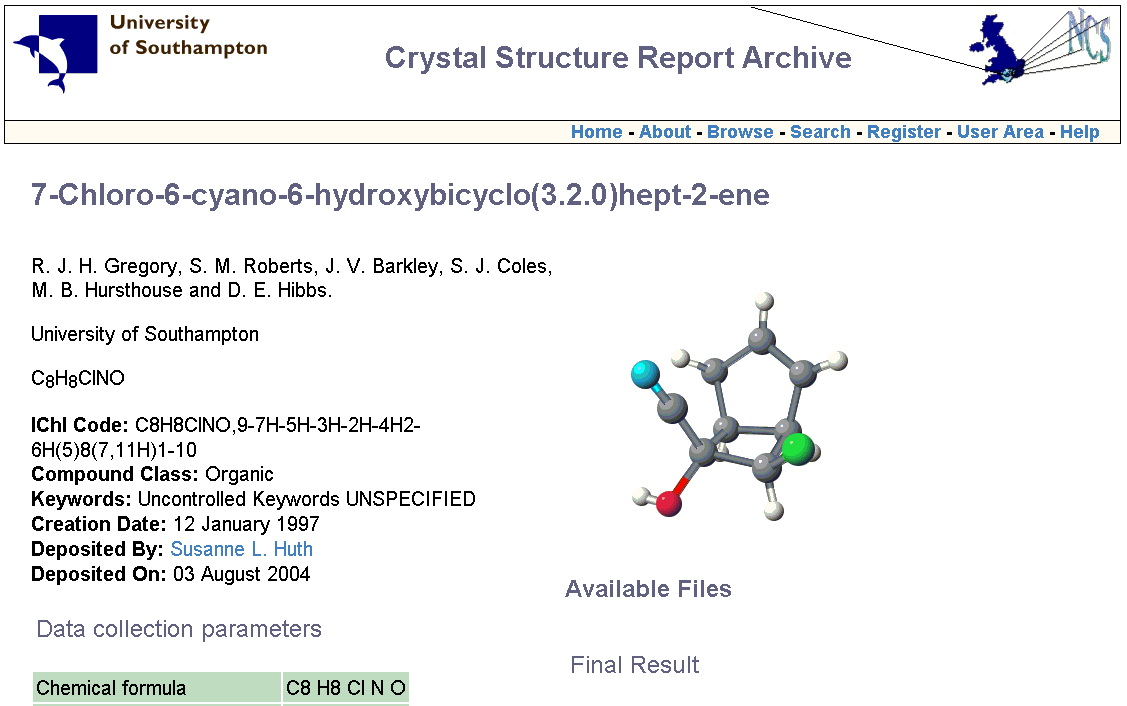
INChI v0.932beta was only available to developers so the number of molecules is known and finite. (We stress that this version is now obsolete and should not be used except for reproducing the current results.) We collaborated with the UK National Crystallographic Service at Southampton and they have published about 72 molecules which include INChIs. We have published about 10 of the INChIs for the NCI dataset. We therefore have a closed world in which the precision and recall can be explored and we hope to be able to provide more details at the meeting
INChIs are generated by the batch program inchi.exe developed at NIST (see other paper). We have wrapped this in a Java graphical interface and also provided it as a Web Service for a variety of common chemical formats. The result is an XML document, containing the INChI String:
 |
 |
Generated INChI:If you have any question, please contact Yong Zhang(yz237@cam.ac.uk). |
|
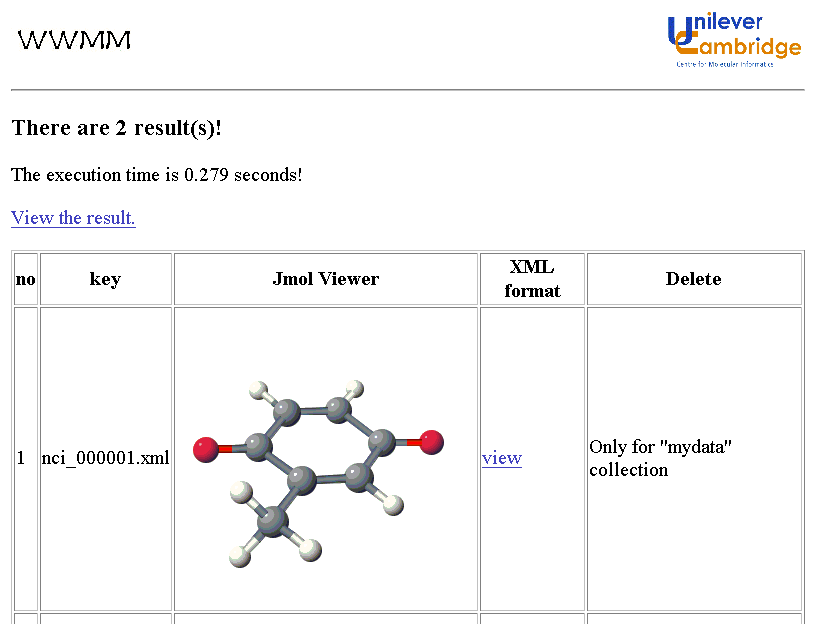
We have customised this into a graphical search query that transmits the INChI directly to the Google API. This system then allows a scientist to draw a molecule and search the complete Web by INChI. A demonstration system is given at: http://wwmm.ch.cam.ac.uk/sdf2inchi/marvin_google.html and some snapshots are given. Note that you MUST select V0.932beta. The Marvin applet is currently convenient but in principle any graphical chemical editor generating CML can be used and we hope to replace it by an OpenSource tool such as JChempaint.
These examples are publicly accessible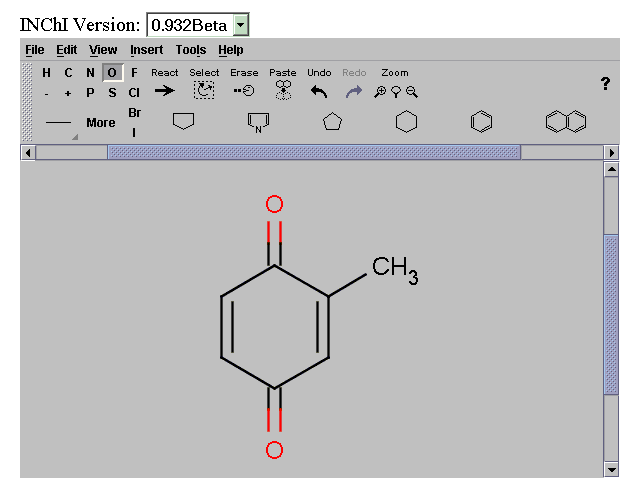
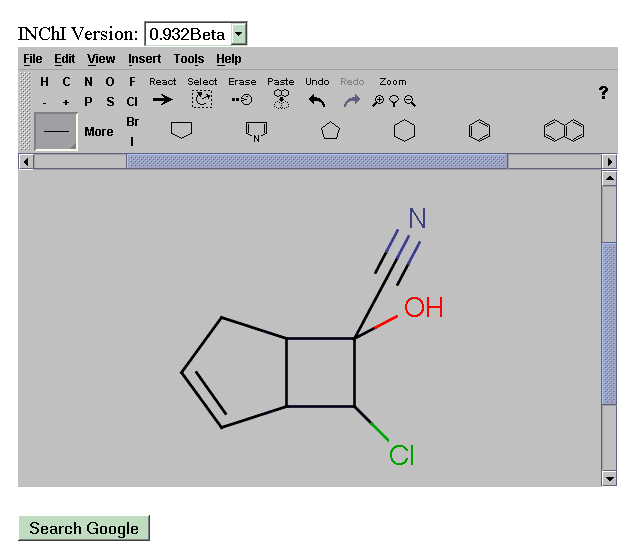
The INChI specification is now stable and only 1.12beta or above should be used. We have now converted the whole KEGG database to INChI1.12 and mounted these on our site. We submitted these to Google on 2004-10-05 and if we are fortunate these may have been indexed by the time of the SWLS meeting, in which case we shall have a meaningful lifeSciences demo.
CML and INChI have been closely correlated and CML is currently the only non-graphical format that INChI can accept. We therefore recommend a combination of INChI and CML as a means of populating the chemical semantic web. We hope that bioscientists will be among the early adopters and will be delighted to collaborate on its development.
This is a compliant XHTML hyperdocument (i.e. a collection of linked components) and we urge the adoption of this technology rather than proprietary formats such as DOC and PDF. It is far easier for machines to extract information from XML than binary formats. We also urge the W3C and other organisations to promote the use of XHTML over PDF and DOC in scientific communication.