- From: M. Scott Marshall <marshall@science.uva.nl>
- Date: Mon, 06 Mar 2006 13:15:07 +0100
- To: chris mungall <cjm@fruitfly.org>
- CC: public-semweb-lifesci@w3.org
- Message-ID: <440C27CB.6030401@science.uva.nl>
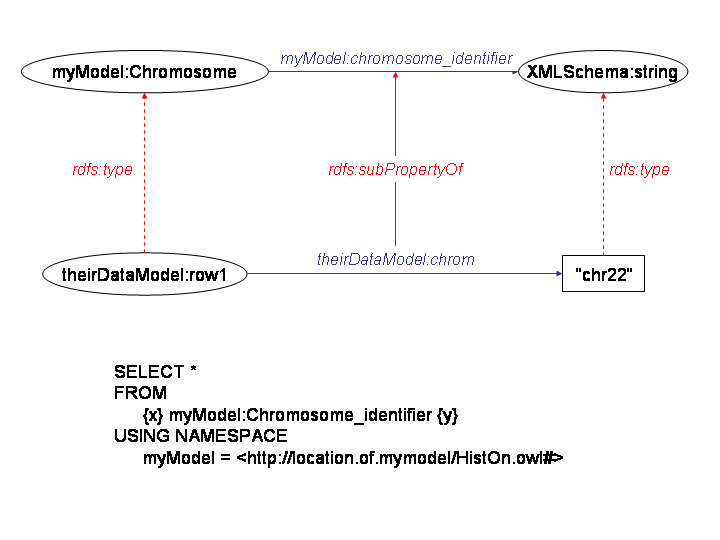
chris mungall wrote: > I think both approaches are a little too XML-centric; fine for a few > use cases but in general the syntax obscures the declarative semantics > of the mapping which must be kept as perspicuous as possible. Why not > just use an RDF query language? This could be used to query over > generic DOM-as-RDF or relations-as-RDF mappings. This would seem to > leave open plenty of room for optimisation, scalability - and the data > sources could be queried directly (albeit slowly) rather than having to > refresh your data warehouse. > > See also > http://scholar.google.com/url?sa=U&q=http://www.ics.forth.gr/isl/ > publications/paperlink/Koffina.pdf Chris - interesting link. As I mentioned earlier[1], I think that describing mappings in a SW language is preferable, if for no other reason than for uniformity. I agree that if you are going to perform the transformation from semantic-less data in RDF (syntactic data types only, with, say, column names from tabular data) to semantically tagged data (the same data but with OWL types), you might as well do it in a RDF query language (preferably SPARQL). We were about to do just that with a SeRQL construct statement but found that we could avoid rebuilding the entire RDF graph of the data by simply declaring 'data-level' properties to be subproperties of OWL types in RDFS (allowing the auto-inferencing of the Sesame RDFS repository to take care of rdfs:type). We were subsequently able to issue data integration queries in our own OWL terms [see attached image]. This seems to work well for "equivalence" mappings. Is there a best practice for this? I think that there are some features of translation to RDF that should be carefully noted. In particular, I think that it is important to distinguish between translations that are direct mappings, where terms are simply translated to RDF syntax ala the YeastHub article[2] and those that include semantic mappings, such as the RDFS subproperty "trick" mentioned above. I agree with Peter Mork where he pointed out in [3] that the road to semantic annotation is best achieved in two explicit transformations: syntactic and semantic. -scott [1]http://lists.w3.org/Archives/Public/public-semweb-lifesci/2006Feb/0171.html [2]http://bioinformatics.oxfordjournals.org/cgi/reprint/21/suppl_1/i85 [3]http://lists.w3.org/Archives/Public/public-semweb-lifesci/2006Feb/0172.html
Attachments
- image/png attachment: subpropertymapping.png
Received on Monday, 6 March 2006 12:15:25 UTC